More Information
Submitted: July 16, 2021 | Approved: August 06, 2021 | Published: August 09, 2021
How to cite this article: Nikhra V. SARS-CoV-2 Infection, COVID-19, and long covid: Saga of erratic immune response, waning immunity, and immune system failure. J Pulmonol Respir Res. 2021; 5: 078-087.
DOI: 10.29328/journal.jprr.1001030
Copyright License: © 2021 Nikhra V. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords:COVID immune landscape; Convergent mutations; D614G G variant; Erratic and overactive response; Immune escape; Intra-host diversity; Variants of concern (VOCs); Variants of interest (VOIs); Vaccine upgradation
SARS-CoV-2 Infection, COVID-19, and long covid: Saga of erratic immune response, waning immunity, and immune system failure
Vinod Nikhra*
Department of Medicine, Hindu Rao Hospital & NDMC Medical College, New Delhi, India
*Address for Correspondence: Dr. Vinod Nikhra, Senior Consultant and Faculty, Department of Medicine, Hindu Rao Hospital and NDMC Medical College, New Delhi, India, Email: [email protected]; drvinodnikhra@ rediffmail.com
Introduction - evolution of SARS-CoV-2 variants: With the unrestrained pandemic for over last one-and-half year, SARS-CoV-2 seems to have adapted to its habitat, the human host, through mutations that facilitate its replication and transmission. The G variant incorporating D614G mutation, potently more transmissible than the ancestral virus arose during January 2020 and spread widely. Since then, various SARS-CoV-2 variants of concern (VOCs) and variants of interest (VOIs) with higher infectivity or virulence or both, have evolved on the background of G variant, and spread widely.
SARS-CoV-2 infection and the immunodynamics: As the virus becomes more transmissible, its lethality may drop. Apart from the humoral immunity, T-cell recognition from a previous SARS-CoV-2 infection or vaccination may modify the disease transmission correlates and its clinical manifestations. On the other hand, the immunity generated may reduce probability of re-infection as well as limit evolution of adaptive mutations, and emergence of highly infectious and immune-escape variants. There are complex issues related to the SARS-CoV-2 evolutionary dynamics and host’s immunodynamics.
Trending etiopathoimmunological correlates: The evolution potential of SARS-CoV-2 is limited because of proofreading function of nsp14. The S protein mutations affect transmissibility, virulence, and vaccine efficacy. The D614G mutation in G variant with higher infectivity has turned the Chinese epidemic into a pandemic. Other SARS-CoV-2 variants, such as Alpha, Beta, Gamma, and Delta seem to have evolved as result of adaptation to selective pressures during periods of prolonged infections and subsequent transmission. Further, there is issue of convergent association of mutations.
Basics of immunity and immune system failure: The nature of the immune response after natural SARS-CoV-2 infection is variable and diverse. There are pre-existing neutralizing antibodies and sensitized T cells elicited during previous infection with seasonal CoVs influencing the disease susceptibility and course. The virus has evolved adaptive mechanisms to reduce its exposure to IFN-I and there are issues related to erratic and overactive immune response. The altered neutralizing epitopes in the S protein in SARS-CoV-2 variants modify the immune landscapes and clinical manifestations.
Conclusion: current scenarios and prospects: Presently, the SARS-CoV-2 infection is widespread with multiple evolving infectious variants. There is probability of its transition from epidemic to endemic phase in due course manifesting as a mild disease especially in the younger population. Conversely, the pandemic may continue with enhanced disease severity due to evolving variants, expanded infection pool, and changing immunity landscape. There is need to plan for the transition and continued circulation of the virus during the endemic phase or continuing pandemic for indefinite period.
Emerging SARS-CoV-2 variants
The Pandemic and Evolution of D614G or G Variant: The mutation process is a natural characteristic of the lifespan of a virus. Over time, the viruses replicate and mutate into more transmissible but less virulent forms to persist inside the host and extend their lifespan. Apparently, the transmissibility and virulence may have an inversely proportional relationship, and in general, the viral mutations lead to higher transmissibility and lower virulence. In other words, with ongoing evolution a virus tends to become more transmissible but less lethal. A particular variant may have higher transmission potential if the infected hosts are shedding more virus. The natural selection appears to act on variation in viral transmission potential and not variation in virulence, per se. There is a probability, though, the virus may evolve to become more virulent, causing more severe disease and host mortality. However, a more severe infection may reduce contact rates of infected individuals, limiting the opportunity for viral transmission.
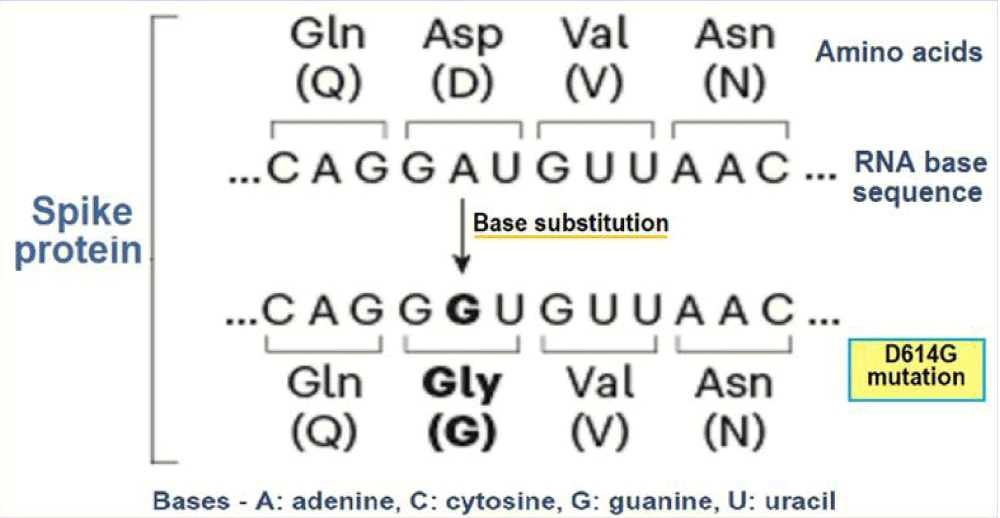
Over last one-and-half year, SARS-CoV-2 seems to have adapted to its habitat, the human host, through mutations that facilitate its replication and transmission [1]. One widespread and more transmissible variant than the ancestral virus is D614G or G variant in which aspartic acid (D) had been replaced with glycine (G) at amino acid position 614 in the S protein (Figure 1).
Figure 1: The evolution of D614G or the G variant.
There is likelihood that the G variant arose during January 2020 in early phase of epidemic in China and became dominant worldwide. The D-to-G substitution appears to have resulted in more efficient infection, replication, and enhanced transmission. The observed increased frequency of infection with SARS-CoV-2 G variant is consistent with the selective advantage for the virus. Further, the D614G form is associated with higher viral loads and younger patient age, though there is no association of with increased severity of infection [2]. In fact, the S protein is made up of three smaller peptides which bind to the ACE2 receptors. The peptides are in open or closed orientation, with the open orientation facilitating their binding with the receptors. The D614 G mutation seems to relax the connexions between the peptides and favouring the receptor bonding and cellular internalization.
The D614G mutation in the S glycoprotein of SARS-CoV-2 was first identified in virus sequences in several Chinese provinces in late January, and later detected in early March 2020 at other places. By June 2020, the G variant replaced the ancestral virus to become the dominant form globally [3]. Further, the mutation seems to have arisen independently and simultaneously across multiple geographic regions, which is suggestive of natural selection and an adaptive benefit of D614G for the virus. As observed, the mutation leads to increased infectivity and transmission than the ancestral virus with 614D but neither causes a more severe illness nor impacts the effectiveness of lab tests, therapies, and vaccines [2]. The G variant, which originally emerged in China, has subsequently spread around the world. The D614G’s spike protein sequence has been used to develop the vaccines in use presently.
Going through the progression of the SAR-CoV-2 infection, it originally started with the L strain that appeared in Wuhan in December 2019. Its first mutation, the S strain appeared at the beginning of 2020, which mutated to G strain by mid-January 2020. The G strain has mutated further into sub-strains GR and GH at the end of February 2020. Presently, the G, GR and GH forms are by far the most widespread and encompass for about 74% of all gene sequences henceforth analysed [4].
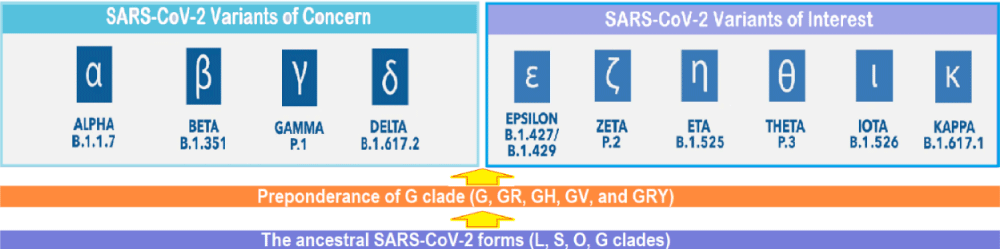
The ongoing evolution of SARS-CoV-2 virus: Various SARS-CoV-2 variants of concern (VOCs) have evolved on the background of D614G mutation which arose during January 2020 and spread widely replacing the ancestral virus. During September-October 2020, a new SARS-CoV-2 lineage, B.1.1.7 (Alpha variant), having substantial fitness advantages over other circulating lineages, rapidly spread from the UK to various countries around the globe. More recently, the SARS-CoV-2 lineage, B.1.617.2 was first detected in India in late 2020 (named Delta variant by WHO later on 31 May 2021), has led to widespread resurgence, and subsequently has spread to other countries. The Delta variant has mutations such as the substitutions T478K, P681R and L452R in the S protein, which lead to higher transmissibility and reduced neutralization by antibodies for previously circulating variants of the COVID-19 virus [5]. Further, its secondary attack rates are stated to be 51% - 67% higher than the Alpha variant, apart from the fatality rate being about 1.9% higher than the Alpha variant [6]. Other VOCs are Beta (B.1.351), Gamma (P.1), and Alpha (B.1.1.7) with E484K. In addition, there are certain variants of interest, such as Epsilon, Zeta, Eta, Theta, Iota, and Kappa. The virus SARS-CoV-2, thus, has the capacity to evolve into more efficient variants (Figure 2).
Figure 2: The ongoing evolution of SARS-CoV-2: From L, S, O and G clades to G, GR, GH, and other G clades, and further evolution from various G clades to Variants of Concern (VOCs) and Variants of Interest (VOIs).
As SARS-CoV-2 continues to spread, the new variants are developing under evolutionary pressures such as host immunity as the result of infection or vaccination, chronicity of SARS-CoV-2 infection, partial and erratic immunity, and perhaps genetic variations in various races and human population groups. There is emerging evidence to suggest that some SARS-CoV-2 variants may carry enhanced antigenicity, leading to likely build-up of differential herd immunity among population groups through natural infection or vaccination. On the other hand, the emergent SARS-CoV-2 lineages may potentially escape vaccine or natural immunity. Additionally, carrier state for indefinite period in those exposed to infection and animal reservoirs may develop to complicate the dynamics of SARS-CoV-2 evolution and adaptation.
SARS-CoV-2 and host immunodynamics
Viral evolution, transmissibility and virulence: In general, as the virus becomes more transmissible, its lethality may drop. Further, there is evidence to show that the T-cell recognition from a previous SARS-CoV-2 infection or vaccination may be able to deal partially with the new variants in terms of modified disease transmission and clinical manifestations. Thus, it is being presumed that the neutralizing effect of T-cells within the population as well as the diminishing lethality of the virus may lower down the infection rate in due course. There are reasons to believe that intermediate levels of immunity generated through natural infection or vaccination may reduce the probability of infection as well as limit the evolution of adaptive mutations by restricting the viral population size within vaccinated hosts. Furthermore, viral loads are lower in infections 12 to 28 days after a single dose of vaccine than in unvaccinated individuals, indicating diminished likelihood of transmission by vaccinated individuals [7]. Similarly, the vaccination is likely to slow the viral evolution. Thus, reducing numbers of infections through vaccination could reduce the chances for variants to be generated, selected, and transmitted [8]. In fact, there is likelihood of emergence rate of immune-escape variants being mitigated by reduced opportunities of the confluence of mutation, selection, and transmission following widespread vaccination.
But the issues related to evolutionary dynamics of SARS-CoV-2 as well as host’s immunodynamics are intricate and full of uncertainties. There is possibility that the SARS-CoV-2 vaccine dosing regimens generating intermediate levels of immunity could accelerate the emergence of new variants, especially the immune-escape variants, capable of escaping immunity wholly or partially induced by prior infection or vaccination [9]. In fact, a noteworthy hypothesis being advanced asserts that such variants may arise through de novo mutation and selection in partially immune hosts due to the weak immune response following infection or vaccination. This conjecture is being challenged as the transmission of SARS-CoV-2 virus typically takes place in the early stage of infection following relatively few cycles of replication [10]. Further, there may not be adequate prospects for adaptive mutants to be generated in a frequency that may lead to onward transmission, and the intermediate levels of immunity may also sufficiently restrict viral replication and thus limit adaptive mutations [11].
It has been observed that those infected with high viral load generally have one or few within-host variants. In other words, in most of SARS-CoV-2 infection, there is low levels of within-host diversity when viral loads are high. Further, the major variant is typically transmitted from the host, whereas the minor variants lost. Only sporadically, the minor variant is transmitted, or multiple variants are transmitted, leading to a mixed infection from SARS-CoV-2 variants. The virulent variant is indicated by severity of the disease as well as the ensuing mortality rate, it can cause. In general, if the virus becomes more transmissible, its lethality may drop. But in this respect, the D614G variant having increased transmissibility, spread globally during the first year of the pandemic with no obvious drop in its virulence. Further, the current VOCs, B.1.1.7 lineage has an estimated transmission advantage of ~50% and B.1.617.2 more transmissible than the former, manifest a higher virulence than the ancestral lineages. In addition, they are presumed to have acquired a decreased sensitivity to natural and/or vaccine-acquired immunity similar to the B.1.351 and P.1 variants, underlining the fact that the evolving SARS-CoV-2 may behave in a complex way with new mutations or combinations of mutations conferring selective advantages to the virus.
Viral phylogenics and epidemiological dynamics: Through identifying SNPs, the viral phylogenics can be used to evaluate viral emergence, characterize the geographical spread, reconstruct epidemiological dynamics, and identify instances of adaptation. As estimated, the SARS-CoV-2 virus evolves at a rate of ∼1.1 × 10−3 substitutions per site per year, which corresponds to one substitution every ∼11 days, and has the most recent common ancestor (TMRCA) around late November 2019 [12]. The phylodynamic analyses can also give information about the virus spread, both spatially and temporally, and allow to identify the viruses circulating in a region as well as new virus introductions. In addition, the phylodynamic techniques may indicate the rate of viral spread through a host population and identify occurrence of viral adaptations. The number of available SARS-CoV-2 genomic sequences is enormous. These are often reported without exact sampling location data because of certain privacy issues. However, viral dynamics may be heterogeneous even within and between geographically close locations. Furthermore, the travel related issues complicate the phylodynamic data. Despite these challenges, a huge reliable SARS-CoV-2 sequence data is available on GISAID’s EpiCov database and on other platforms such as Nextstrain and Microreact. Because of their potential phenotypic effects, evolution of genomic insertions, deletions, and recombinants are of paramount significance and need to be monitored.
The viruses evolve as a result of mutations and natural selection for favourable traits such as more efficient viral replication, transmission, and evasion of host defences. In general, the viral adaptations take place through the evolution of novel viral traits such as immune escape through genetic variation. The latter occurs through nucleotide substitutions, misincorporations, insertions or deletions. Apart from this, the recombinations are common in CoVs during replication and give rise to new SARS-CoV-2 lineages. But the diversity of SARS-CoV-2 is limited because of proofreading function of the 3′-5′ exonuclease nsp14. Thus, the vaccines based on a single sequence of the viral S protein are likely to generate an immune response protective to various circulating variants. However, the newer variants of SARS-CoV-2 with mutations in S protein have emerged, posing potential challenges for vaccination and antibody-based therapies. The continuing pandemic and unprecedented spread of SARS-CoV-2 may lead to the possibility for accumulation of additional consequential mutations in S protein and throughout the viral genome [13].
Trending etiopathoimmunological correlates
Intra-host diversity and onward transmission: The SARS-CoV-2 shares high sequence homology with SARS-CoV both in genomic structure and host receptor preference. Whereas SARS-CoV-2 and the common cold human coronavirus, HCoV-NL63, though both recognize ACE2 as the host cell receptor, have major sequence and structural differences in the receptor-binding domain (RBD) of S protein. This diversity indicates that CoVs can potentially tolerate changes in both sequence and structure without substantial loss of function. The S protein has two subunits, S1 which contains an amino (N)-terminal domain (NTD) and receptor-binding domain (RBD), and the subunit S2 which mediates virus–host cell fusion. The antibody-neutralizing epitopes are scattered throughout S protein but are mostly concentrated in the RBD. As such, the evolution potential of SARS-CoV-2 is limited because of proofreading function of nsp14. The understanding about the individual phenotypic effects of the S1 mutations is emerging, and it has been noted that substitutions and deletions in S1 can affect transmissibility (Tr), vaccine efficacy (Ef), and virulence (Vi). The first notable evolutionary event was the D614G (Asp614→Gly) substitution, which increased the ACE2 affinity, leading to higher infectivity and transmissibility of the G variant.
Further, the substitution at position Asn501 with Thr or Phe increases affinity for ACE2 binding [14]. The substitution at position 452, a leucine-to-arginine substitution (L452R), confers higher affinity of the S protein for the ACE2 receptor and decreased recognition by immune system. Whereas the substitution at position 681, a proline-to-arginine substitution (P681R), facilitates cleavage of the S precursor protein to the active S1/S2 configuration and, thus, boosts cell-level infectivity. The variants may have reduced sensitivity to neutralizing antibodies that bind to the RBD because of triple substitutions of key amino acids, Lys417, Glu484, and Asn501, in the RBD at the ACE2-binding interface or the NTD. It is possible that there is a convergent association of mutations. The mutations that reduce neutralizing antibody binding, such as E484K, may require compensatory mutations that restore infectivity, such as N501Y, as is the case in the B.1.351 and P.1 lineages. Similar situation occurs in B.1.1.7[K] or Alpha[K] lineage, where E484K is present with N501Y. The role of compensatory mutations is also supported by the emerging B.1.525 lineage that has both E484K and Δ69–70 leading to reduced antibody sensitivity and compensatory increase in infectivity, respectively.
The diversity of SARS-CoV-2 is limited because of proofreading function of the 3′-5′ exonuclease nsp14. Albeit, the proofreading capacity of nsp14 is limited to point mutations and it cannot repair other greater alterations such as deletion, insertion, recombination or misincorporation. Thus, Significant intra-host evolution of SARS-CoV-2 can occur as reported various case studies in some patients with protracted infection due to impaired immunity [15,16]. These patients had up to fivefold reduced neutralization sensitivity to convalescent plasma (CP) and/or monoclonal antibody therapy, in addition to shedding high titers of SARS-CoV-2 and having active SARS-CoV-2 infection for an average of 115 days before clearing the infection or succumbing to COVID-19 [17,18]. These case studies in immunocompromised patients have documented the deletions of amino acids 69 to 70 (Δ69–70), Δ141–144, or Δ242–248 in S1; the N501T (Asn501→Thr) or N501Y (Asn501→Tyr) mutations; and the E484K (Glu484→Lys) and Q493K (Gln493→Lys) mutations in the RBD. There have been found various deletions in the amino (N)-terminal domain (NTD) of S1 in B.1.1.7 and B.1.351. In addition, other mutations in these variants include K417N, E484K, and N501Y. It is noteworthy that these reports preceded the detection of three major circulating variants—B.1.1.7, B.1.351, and P.1, which contain at least eight single, nonsynonymous nucleotide changes, including E484K, N501Y, and/or K417N (Lys417→Asn) in the RBD [19]. Occasionally, SARS-CoV-2 virus can evolve into multiple distinct lineages within the same infected individual [20].
The complex process of immune response: Several studies suggest that the major circulating variants, such as B.1.351, P.1, and B.1.1.7 lineages, and probably B.1.617.2 lineage have reduced neutralizing sensitivity to convalescent plasma and plasma from recently vaccinated individuals [21,22]. Though, the reduced antibody sensitivity against these variants do not invariably prove that a vaccine is not effective, still the recent data suggests that certain vaccines may be especially less protective against the B.1.351 variant [23]. Further, there is growing concern for the emergence of immune escape mutants in a protracted SARS-CoV-2 infection. Similarly, the partial roll-out and incomplete immunization of individuals leading to suboptimal titers of neutralizing antibody could also promote evolution of escape variants. On the other hand, halting the spread of SARS-CoV-2 through a coordinated and comprehensive vaccination programme and prevention strategies like masking and social distancing are likely to prevent the evolution of immune escape variants.
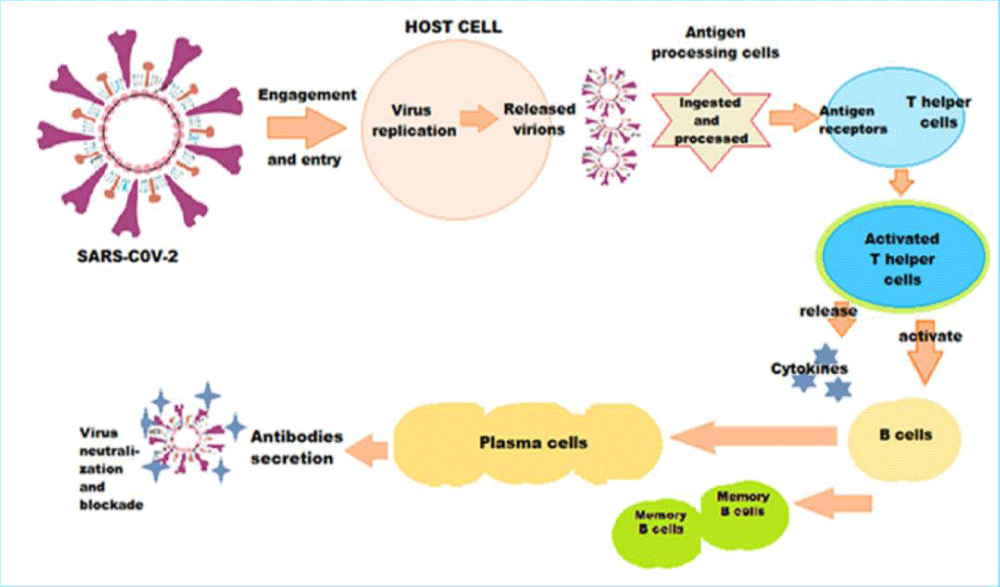
The immune response following SARS-CoV-2 infection or COVID-19 vaccination is a complex process (Figure 3). The antigenic presentation following infection or vaccination activates T helper cells, which in turn activate the B cells. The latter diversify into antibody producing effector B cells and memory B cells. In addition, there are numerous additional infection-induced or vaccine-induced responses pertaining to the innate and adaptive immune system. The responses may protect against infection and further viral immune escape. Conversely, there are uncharacterized mutations outside of S that could facilitate SARS-CoV-2 immune evasion.
Figure 3: The intricate pathophysiology of immune response following SARS-CoV-2 infection or vaccination - The antigenic presentation activates T helper cells, which in turn activate the B cells, which diversify into antibody producing effector B cells and memory B cells. Simultaneously, numerous additional infection-induced or vaccine-induced responses pertaining to the innate and adaptive immunity play significant role.
Genomic variations and phenomenal concerns: The SARS-CoV-2 genomic variants have been emerging and circulating around the world throughout the COVID-19 pandemic. Having evolved in certain geographical regions, they have spread to other regions. Some of them are Variants of Concern (VOC) and Variants of Interest (VOI) labelled by WHO [24]. In short, a VOC is a SARS-CoV-2 variant with demonstrable increase in transmissibility, increase in virulence or a significant change in clinical presentation, or decrease in effectiveness of public health measures or available diagnostics, vaccines, therapeutics. Whereas a VOI is a SARS-CoV-2 variant with genomic changes affecting transmissibility, disease severity, immune escape, diagnostic or therapeutic escape, and ability to enhance community transmission resulting in multiple COVID-19 clusters at multiple geographical locations with increasing number of cases or having other epidemiological impacts to leading to an emerging risk to global public health.
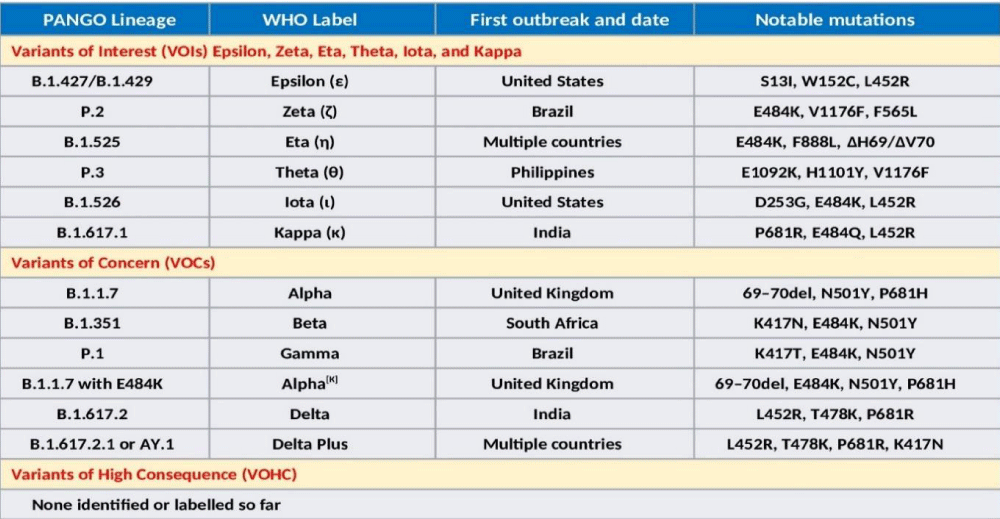
Presently, VOCs include Alpha (B.1.1.7 – UK), Alpha[K] (B.1.1.7 with E484K), Beta (B.1.351 – SA), Gamma (P.1 – Brazil), and Delta (B.1.617.2 – India). In addition, there are VOIs, which include Epsilon (B.1.427/B.1.429 – US), Zeta (P.2 – Brazil), Eta (B.1.525 – multiple countries), Theta (P.3 – Philippines), Iota (B.1.526), and Kappa (B.1.617.1 – India). The lists are likely to be larger in near future. Simultaneously, more infectious variants will keep on replacing those less infectious throughout the world regions and population groups. The WHO has declared Epsilon (1 March 2021 – California, the US), Zeta (17 March 2021 - Brazil), Eta (17 March 2021 – originated in Nigeria, now in multiple countries), Theta (24 March 2021 - Philippines), Iota (24 March 2021 – New York, the US), and Kappa (4 April 2021 - India) as VOIs. Another variant, A.23.1 with altered spike, has emerged and is responsible for Uganda epidemic [25].
It is likely that the VOIs and VOCs are the result of selective pressures and adaptation of the virus during periods of prolonged infections and subsequent transmission. Further, there is a convergent association of mutations as well. Potentially, such variants containing new mutations will continue to emerge in different geographic locations as the result of intra-host selection and subsequent transmission. The SARS-CoV-2 VOCs and VOIs cause a significant increase in transmissibility or virulence or both. Further, it has been speculated that SARS-CoV-2 may continue to accumulate mutations that evade immune responses. The major SARS-CoV-2 Variants have been classified by the Centers for Disease Control and Prevention (CDC) into 3 broad categories, VOIs - Variants of Interest, VOCs - Variants of Concern, and VHCs - Variants of High Consequence (26). The classification relates the etiopathological correlates to the infectivity and virulence of the emerging variants (Figure 4).
Figure 4: The Variants of Concern (VOCs) and Variants of Interest (VOIs), the WHO nomenclature, geographical evolution, and notable mutations.
The B.1.1.7, also called 501Y.V1 variant, emerged in the UK, during September 2020. It is significantly more infectious and has 18 mutations including 9 in the S gene and others in ORF1ab/ORF8/N. It has N501Y mutation in S affecting bonding to the human ACE2 receptor and being considered more virulent than D614G, able to cause severe disease in a higher proportion of patients. The current vaccines appear to fully protect against B.1.1.7 variant.
The B.1.351, which originally emerged in South Africa, has 8 mutations, 5 in S including the N501Y mutation found in the B.1.1.7 variant, and others in ORF1ab/E/N. The B.1.351 (501Y.V2) variant carries additional immune evasion mutations, notably the E484K (Glu484→Lys) mutation and appears to not well-recognized by the immune systems of people previously infected with D614G. The variant shows substantial reduced neutralizing activity of therapeutic monoclonal antibodies (mAbs). Further, the vaccine efficacy is not strong to this variant.
The Gamma variant, P.1 carries 21 mutations including 10 in S including N501Y, others are in ORF1ab/ORF3a/ORF 8/ORF9/ORF 14/N. In short, it incorporates E484K on the B.1.1.7 background. Given that the N501Y (Asn501→Tyr) mutation arose spontaneously in Alpha, Beta, and Gamma variants, it appears to confer a competitive advantage for the SARS-CoV-2 variants by increasing the affinity of spike for ACE2 and together with other less well characterized mutations has resulted in enhanced infectivity.
The Delta (B.1.617.2) genome has 13 mutations, 15 or 17 according to some other sources. The three of them, L452R, T478K, P681R, present the spike protein are of particular concern. The Delta Plus variant is supposed to be formed due to a mutation called K417N in the Delta or B.1.617.2 variant. There is exchange at position 417 (lysine-to-asparagine substitution) in Delta plus variant. The K417N mutation in S protein is associated with immune escape and reduced susceptibility to vaccine, and mAbs therapy. Globally, more than 12 countries have detected Delta Plus cases.
Basics of immunity and immune system failure
Imperfect humoral, T-cell, and interferon response: The infection with seasonal and cross-reacting CoVs is common in humans. In general, the CoVs do not provoke a fully protective immunity, and repeat infections are common. The studies show that detectable antibody levels wane over the first few months, post-infection [27]. This is exemplified by the ineffective serum antibody levels against the seasonal human coronavirus OC43 - HCoV-OC43 [28]. Similarly, the vaccines tend to be non-efficient at preventing seasonal CoV infection [29]. But the HCoV-specific immunity may wane but is not lost. In fact, they are less efficient than natural infections at provoking immunity and carry risks of adverse cross-reactions. As related to the SARS-CoV-1 infection, the humoral immunity may last up to 2 to 3 years, but antigen-specific T cells against the virus have been detected 11 years after infection [30].
The nature of the immune response after natural SARS-CoV-2 infection is complex and diverse. There are intricacies in COVID-19 dynamics and immunological characterization to SARS-CoV-2 infections. There is evidence of pre-existing T cells and antibodies capable of cross-reacting with SARS-CoV-2 suggests that immunological memory responses elicited during infection with seasonal coronaviruses may also affect COVID-19 susceptibility and disease risk as well as clinical manifestations. In this context, it has been hypothesized that severe COVID-19 manifestations may arise due to presence of non-neutralizing antibodies from prior CoV infections [31]. Further, the T cell–mediated response is likely to play an important role in regulating the SARS-CoV-2 viral replication and disease manifestations.
There are variations in the immune response to a primary SARS-CoV-2 infection. The individual factors like earlier exposure to a CoV infection as well as the genetic factors may lead to different immune landscapes and affect clinical manifestations and severity of the disease. Considering the common four circulating HCoVs in children and adults which cause common cold and that the primary infections usually occurs early in life, the reinfection with a CoV later may cause a recall response. The pre-existing non-neutralizing antibodies from prior CoV infections as well as the T-cell-mediated response dependent on tenacity and recall of immune memory in a population group, thus, may affect the course of the pandemic ranging from recurring outbreaks and resurgence to its near-elimination.
Loss of neutralizing epitopes in the S protein in the SARS-CoV-2 variants is likely to reduce the immune response [32]. In this context, the T cell mediated response may feature as a superior correlate of protection (CoP) against COVID-19 as the new more infectious and virulent variants emerge. The CD4+ and CD8+ T cell response encompasses specificity to several hundred epitopes across the entire SARS-CoV-2 proteome, the majority of which are unpaired in the virus. The T cell epitopes that are altered in the SARS-CoV-2 variants are likely to bind to the various human leukocyte antigen (HLA) molecules presenting antigenic peptides to T cells, even when binding affinities are altered. Thus, the variants, such as Alpha variant, disabling the first line of immune defense, get more time to multiply, and may overcome the T cell mediated response.
There exists another line of anti-viral defence, represented by production and secretion of interferon by the host cells. But like many other viruses, SARS-CoV-2 has evolved mechanisms to reduce its exposure to IFN-I [33]. Its Alpha variant drives down the production of interferon by infected lung cells by flooding with Orf9b proteins. Whereas the Beta and Delta variants drive down interferon production in the infected cells through a different mechanism. The nsp13, nsp14, nsp15, and orf6, orf8, and the M protein are potent inhibitors of the MAVS pathway, leading to inhibition of IFNβ production. The severe COVID-19 is associated with exhaustion of CD4+ and CD8+ T cells, as a result of deficient IFN-I production. Further, the IFN-I production, is significantly impaired in obesity and metabolic syndrome, and with ageing.
The erratic and overactive immune reactions: The innate immunity is the first line of defence against the SARS-CoV-2 virus invasion. The recognition of pathogen results in subsequent cytolytic immune responses, mainly through the type I interferons (IFN) and natural killer cells. The adaptive immunity also plays an important part in viral clearance via activated cytotoxic T cells that destroy virus-infected cells and the antibody-producing B cells target virus-specific antigens. The anti-viral immune response is crucial to eliminate the invading virus, but an overactive and persistent anti-viral immune response may lead to massive production of inflammatory cytokines and damage the host tissues. One of the major causes of ARDS in COVID-19 patients is a hyperactive immune system. The overproduction of cytokines caused by aberrant immune activation is known as a cytokine storm. The alveolar macrophages expressing ACE2 are a prime target cells for SARS-CoV-2 infection and these activated macrophages may play an important role in the hyperinflammatory syndrome due an overactive Immune response [34].
There is a complex immune landscape generated by SARS-CoV-2 infection [35]. The issue of immunity with SARS-CoV-2 infection is complex and COVID-19 as a disease weakens the immune system as well as leads it to act in erratic manner leading to the cytokine storm in certain cases. The immune efficacy with respect to susceptibility does not appear to prevent reinfection, but may attenuate the severity, possibly retard reinfection and may or may not reduce transmissibility or infectiousness. There is possibility that symptoms due to a CoV reinfection may be mild, and the virus may be cleared more quickly. The rapid rise in both IgM and IgG following a CoV infection, indicates that earlier primary infection with a common endemic HCoV strain provokes a recall response and the resurged HCoV-specific immunity may influence the course of COVID-19 illness.
The viral variant and other pathogen characteristics also have impact on the course of COVID-19 infection and disease. Patients with severe disease have substantially lower lymphocyte counts with reduced number of CD4+ T cells, CD8+ T cells, and natural killer cells. Whereas the proinflammatory subsets of T cells, including IL-17-producing CCR4+ CCR6+ CD4+ (T-helper 17 or Th17) cells and perforin and granulysin-expressing cytotoxic T cells are increased. It has been indicated that the particular helper T cell population, tissue-resident memory-like Th17 cells (Trm17), in the lungs of patients with severe COVID-19 may be central to the development of hyperinflammation, lung injury, and subsequent ARDS. In addition, there are present high plasma concentrations of inflammatory cytokines such as IL-6 and tumor necrosis factor (TNF). The Trm17 cells may become activated as part of a cytokine storm, during which they start producing inflammatory molecules like GM-CSF [36].
Various clinical reports suggest that a subgroup of severely affected patients exhibit a hyper-inflammatory response to COVID-19 (COV-HI). The cytokine storm is associated with COV-Hi and organ damage in severe COVID-19. The exaggerated production of inflammatory cytokines including TNF-α and IFN-γ leads to inflammatory cell death and PANoptosis, characterized by gasdermin-mediated pyroptosis, caspase-8-mediated apoptosis, and MLKL-mediated necroptosis [37]. This group of patients has a higher plasma concentration of IL-2, IL-7, IL-10, granulocyte-colony stimulating factor, IFNγ-induced protein-10 (IP-10), macrophage chemoattractant protein-1, macrophage inflammatory protein 1α, and TNF. There may be hypercytokinaemia, unremitting fever, cytopenias, hyperferritinemia, and multi-organ damage, in these severely ill patients.
Immune response to COVID-19 vaccines: The present vaccines available for COVID-19 prophylaxis are based on the wild-type viral S protein. Most vaccinated people develop neutralizing antibody (Ab) with an IC50 (half maximal inhibitory concentration) within the protective margin, although precise correlates of protection (CoP) are unknown. Variants with E484K mutations and other escape mutants may bring down vaccine efficacy, prompting the need for new and updated vaccines [38]. Further, there is a complex immune landscape generated by vaccination program in face of prevalence of various SARS-CoV-2 variants as the population groups may comprise of non-exposed, exposed, and non-infected, and exposed and infected persons with history of asymptomatic or mild disease and those with moderate to severe disease. Further, the individual immune response and the cumulative immune landscape of the population are important for SARS-CoV-2 primary and secondary infection and vaccination and determine the prospect for the pandemic.
As the pandemic is evolving and spreading to newer geographical regions, various genetic, racial, geographical, and socioeconomical factors are bound to influence the course of the pandemic. Various analyses consider that the severity of infection with SARS-CoV-2 may change in due course over a span of years in future. The epidemiological and immunological data indicate that the infection-blocking immunity for the virus wanes rapidly, but that disease-reducing immunity is long-lived [39]. Further, there is likely to be transition from epidemic to endemic dynamics associated with a shift in the age distribution of primary infections to younger age groups. Here, an optimistic model envisages that once the endemic phase is reached and primary exposure is in childhood, the virulence of SARS-CoV-2 may go down. Further, depending on its immune response, a vaccine could accelerate the state of mild disease endemicity by cultivating herd immunity. However, there might be a different outcome for the emergent infection causing a severe disease in children. Both situations reinforce the importance of vaccination and behavioral practices for disease containment.
There is question of the functional immunity to reinfection, disease, and carrier state with viral shedding and the endemicity of SARS-CoV-2 in the long run. The longitudinal analysis of SARS patients provides an opportunity to measure the durability of immune memory in the absence of re-exposure. In contrast to the antibodies, the memory T cells persist for much longer periods in animal models, and the immunity induced by previous strains is potent to prevent severe disease. However, the effect of genomic variation may influence the vaccine-induced immunity in light of the narrow epitope repertoire of currently available vaccines. On the other hand, the immune evasion is supposed to come at a biological fitness cost to the virus and impose an upper limit to the number of probable mutations when faced with the neutralizing antibody repertoire [40]. The similar mutations arising recurrently in the spike through convergent evolution in geographically distinct isolates also indicate that the spike variants offering a survival advantage to the virus are limited.
It appears that frequent and regular boosting of immunity by repeat doses of the COVID-19 vaccine may be required to maintain protection from the virus especially in light of temporary and waning immunity. Apparently, mimicking the natural immunity by vaccination may be a practical prophylactic strategy. Thus, during the transition of pandemic to endemicity, the SARS-CoV-2 infections may frequently occur in older individuals, and the immunity induced by infection or vaccination similar to that produced by natural infections in childhood may be a desirable outcome. Further, if the vaccine is able to cause a major reduction in transmission, in long run it may be suitable to consider strategies to vaccinate the older individuals for whom infection can cause a severe morbidity and higher mortality, while allowing natural immunity and transmission to be maintained in younger individuals.
Current scenario and prospects
Unrestrained pandemic and future scenario: Using symptoms as a surveillance tool to curb the spread of SARS-CoV-2 is difficult, as milder reinfections increasingly contribute to ongoing transmissions [41]. Further, the infection or vaccination may protect against disease but may not provide transmission-blocking immunity needed for achieving significant long-term herd immunity. Furthermore, social distancing, masking, and an effective vaccination are critical for control of the pandemic and during its transition to endemicity. Once the endemic phase is reached and primary cases during childhood may manifest as mild disease, mass vaccination may no longer be necessary. But, if the primary infections in children are severe, then vaccination during childhood will need to be continued. Further, in light of the possible changes in the disease severity due to evolving variants and changing immunity landscape, we need to plan carefully for the transition to endemicity and the ongoing circulation of the virus.
Presently, the SARS-CoV-2 infection is so widespread with multiple evolving infectious variants that the hope of its elimination is bleak. However, there can be envisaged a possible outcome that the acquired immunity from infection, reinfection and vaccination may lead to its endemicity [32]. As such, the future trajectory of the disease may be difficult to predict in a set of non-cohesive population comprising of various age groups, harbouring different risk factors, and having varied background in matter of exposure to the infection, manifestation of the disease, and inadequate immunization [42]. Further, though within-host emergence of escape mutants seems to be uncommon during early infection when viral loads are high, occurrence of immune-escape variants in high-viral-load samples emphasizes caution and need for continued vigilance [43].
The watchful and optimistic approach: The laboratory studies test the vaccine efficacy with a focus on level of the antibodies and their ability to block the virus from infecting cells, the immune response to the infecting virus is complex. In the human body the antibodies are a part of the immune response supplemented by the immune T cells, which help in curtailing the infection by identifying and eliminating the infected cells and help to protect from evolving a mild illness to severe disease [44]. Several COVID-19 vaccines appear to work against the VOCs and VOIs, as well. The earlier reports suggested that the vaccines might not work against of the variants, such as the Beta (B.1.351) variant, but the real-world data out of Qatar suggests that the Pfizer vaccine works satisfactorily well against it [45].
The immune response against the COVID-19 vaccine, in general, is robust. The full vaccination seems to offer 75% protection against B.1.351 infections, less than the 95% efficacy reported in the trials but still to a significant level. Simultaneously, there is evidence from clinical studies that the T-cell response may provide significant protection against emerging SARS-CoV-2 variants, as well [46]. There is evidence that when the vaccinated people get infected, are protected from the severe disease and serious outcomes [47]. Compared to this, the role of the vaccine in protecting Long Covid manifestations is being assumed, though not backed by large clinical studies [48].
Upgradation of the COVID-19 vaccines: On entering host cells, the SARS-CoV-2 virus starts replicating. With replication, the chances of random errors, or mutations, crop up during the viral replication cycle, despite the proofreading function of the nsp14 protein. Most of these errors are inconsequential, only a few being of epidemiological concern. Various mutations or their particular combinations, which improve the survival SARS-CoV-2 virus are emerging in different world regions, a phenomenon known as convergent evolution [49]. These particular combinations of mutations are seen to occur over and over again. The first major variant with spike-protein mutation, D614G has helped transmission of SARS-CoV-2 world over. Later, the Alpha (B.1.1.7) variant, has fast replaced those less infectious. The mutation, E484K helps the virus to evade the immune response. In due course, the variants carrying advantageous mutations outcompete a variant that is lacking them and replace it [50].
In fact, the phenomenon of convergent evolution may be a hopeful sign, as the virus may run out of ways to adapt to its habitat. In fact, the convergent evolution may be seen as the game of Tetris, where a limited number of building blocks can be assembled in different but limited combinations [51]. Thus, controlling infections, may hopefully limit the number of evolving variants, and vice versa. There is another aspect of phenomenon of convergent evolution. Faced with new variants, the current vaccines will eventually but gradually become less effective. But, as the virus has a limited number of viable mutations, the vaccines may be updated time to time based on the immunodynamics of evolving SARS-CoV-2 variants to maintain their efficacy. Further, the variant-specific boosters will prompt a more effective immune response against the new variants. Further, mixing vaccines could help boost immunity and help stop variants from bypassing the immune system. The odds are that the newer variants will continue to evolve, hence we have to update and revise our tools and tactics.
- Hou YJ, Chiba S, Halfmann P, Ehre C, Kuroda M, et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020; 370: 1464-1468. PubMed: https://pubmed.ncbi.nlm.nih.gov/33184236/
- Volz E, Hill V, McCrone JT, Price A, Jorgensen D, et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell. 2021; 184: 64-75. PubMed: https://pubmed.ncbi.nlm.nih.gov/33275900/
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, et al. Sheffield COVID-19 Genomics Group. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020; 182: 812-827.e19. PubMed: https://pubmed.ncbi.nlm.nih.gov/32697968/
- Guzzi PH, Mercatelli D, Ceraolo C, Giorgi FM. Master Regulator Analysis of the SARS-CoV-2/Human Interactome. J Clin Med. 2020; 9: 982. PubMed: https://pubmed.ncbi.nlm.nih.gov/32244779/
- Starr TN, Greaney AJ, Dingens AS, Bloom JD. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Reports Medicine. 2021; 2: 100255.
- The Public Health England. https://www.gov.uk/government/publications/investigation-of-sars-cov-2-variants-of-concern-routine-variant-data-update
- Levine-Tiefenbrun M, Yelin I, Katz R, Herzel E, Golan Z, et al. Decreased SARS-CoV-2 viral load following vaccination. Medrxiv Preprint. 2021.
- Cobey S, Larremore DB, Grad YH, Lipsitch M. Concerns about SARS-CoV-2 evolution should not hold back efforts to expand vaccination. Nat Rev Immunol. 2021; 21: 330-335. PubMed: https://pubmed.ncbi.nlm.nih.gov/33795856/
- Saad-Roy CM, Morris SE, Metcalf CJE, Mina MJ, Baker RE, et al. Epidemiological and evolutionary considerations of SARS-CoV-2 vaccine dosing regimes. Science. 2021; 372: 363-370. PubMed: https://pubmed.ncbi.nlm.nih.gov/33688062/
- He X, Lau EHY, Wu P, Deng X, Wang J, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020; 26: 672-675. PubMed: https://pubmed.ncbi.nlm.nih.gov/32296168/
- Hanage WP, Russell CA. Partial immunity and SARS-CoV-2 mutations. Science. 2021; 372: 354. PubMed: https://pubmed.ncbi.nlm.nih.gov/33888632/
- Martin MA, VanInsberghe D, Koelle K. Insights from SARS-CoV-2 sequences. Science. 2021: 371: 466-467. PubMed: https://pubmed.ncbi.nlm.nih.gov/33510015/
- McCormick KD, Jacobs JL, Mellors JW. The emerging plasticity of SARS-CoV-2. Science. 2021; 371: 1306-1308. PubMed: https://pubmed.ncbi.nlm.nih.gov/33766871/
- Starr TN, Greaney AJ, Hilton SK, Ellis D, Crawford KHD, et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 2020; 182: 1295-1310. PubMed: https://pubmed.ncbi.nlm.nih.gov/32841599/
- Avanzato VA, Matson MJ, Seifert SN, Pryce R, Williamson BN, et al. Case Study: Prolonged Infectious SARS-CoV-2 Shedding from an Asymptomatic Immunocompromised Individual with Cancer. Cell. 2020; 183: 1901-1912. PubMed: https://pubmed.ncbi.nlm.nih.gov/33248470/
- Baang JH, Smith C, Mirabelli C, Valesano AL, Manthei DM, et al. Prolonged Severe Acute Respiratory Syndrome Coronavirus 2 Replication in an Immunocompromised Patient. J Infect Dis. 2021; 223: 23-27. PubMed: https://pubmed.ncbi.nlm.nih.gov/33089317/
- Camprubí D, Gaya A, Marcos MA, Martí-Soler H, Soriano A, et al. Persistent replication of SARS-CoV-2 in a severely immunocompromised patient treated with several courses of remdesivir. Int J Infect Dis. 2021; 104: 379-381. PubMed: https://pubmed.ncbi.nlm.nih.gov/33359065/
- Hensley MK, Bain WG, Jacobs J, Nambulli S, Parikh U, et al. Intractable COVID-19 and Prolonged SARS-CoV-2 Replication in a CAR-T-cell Therapy Recipient: A Case Study. Clin Infect Dis. 2021; 73: e815-e821. PubMed: https://pubmed.ncbi.nlm.nih.gov/33507235/
- Kemp SA, Collier DA, Datir RP, Ferreira IATM, Gayed S, et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021; 592: 277-282. PubMed: https://pubmed.ncbi.nlm.nih.gov/33545711/
- Choi B, Choudhary MC, Regan J, Sparks JA, Padera RF, et al. Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. N Engl J Med. 2020; 383: 2291-2293. PubMed: https://pubmed.ncbi.nlm.nih.gov/33176080/
- Liu C, Ginn HM, Dejnirattisai W, Supasa P, Wang B, et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021; 184: 1–17.
- Wang P, Casner RG, Nair MS, Wang M, Yu J, et al. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021; 29: 747–751. PubMed: https://pubmed.ncbi.nlm.nih.gov/33887205/
- Planas D, Bruel T, Grzelak L, Guivel-Benhassine F, Staropoli I, et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med. 2021; 27: 917–924. PubMed: https://pubmed.ncbi.nlm.nih.gov/33772244/
- WHO – Variants of Concern and Variants of Interest. 2021. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants
- Bugembe DL, Phan MVT, Ssewanyana I. A SARS-CoV-2 lineage A variant (A.23.1) with altered spike has emerged and is dominating the current Uganda epidemic. medRixv Preprint. 2021.
- Updated 2021, CDC, USA. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html.
- Self WH, Tenforde MW, Stubblefield WB, Feldstein LR, Steingrub JS, et al. Decline in SARS-CoV-2 Antibodies After Mild Infection Among Frontline Health Care Personnel in a Multistate Hospital Network — 12 States. April–August 2020. MMWR Morb Mortal Wkly Rep. 2020; 69: 1762–1766. PubMed: https://pubmed.ncbi.nlm.nih.gov/33237893/
- Yamaguchi T, Shinagawa T, Kobata H, Nakagawa H. Immunity against seasonal human coronavirus OC43 mitigates fatal deterioration of COVID-19. Int J Infect Dis. 2021; 109: 261-268. PubMed: https://pubmed.ncbi.nlm.nih.gov/34273512/
- Peiris M, Leung GM. What can we expect from first-generation COVID-19 vaccines? The Lancet. 2020; 396: 1467-1469. PubMed: https://pubmed.ncbi.nlm.nih.gov/32971042/
- Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020; 584: 457–462.
- Beretta A, Cranage M, Zipeto D. Is Cross-Reactive Immunity Triggering COVID-19 Immunopathogenesis? Front Immunol. 2020; 11: 567710. PubMed: https://pubmed.ncbi.nlm.nih.gov/33178193/
- Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020; 9: e61312. PubMed: https://pubmed.ncbi.nlm.nih.gov/33112236/
- Sa Ribero M, Jouvenet N, Dreux M, Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020; 16: e1008737. PubMed: https://pubmed.ncbi.nlm.nih.gov/32726355/
- Felsenstein S, Herbert JA, McNamara PS, Hedrich CM. COVID-19: Immunology and treatment options. Clin Immunol. 2020; 215: 108448. PubMed: https://pubmed.ncbi.nlm.nih.gov/32353634/
- Bolouri H, Speake C, Skibinski D, Long SA, Hocking AM, et al. The COVID-19 immune landscape is dynamically and reversibly correlated with disease severity. J Clin Invest. 2021; 131: e143648. PubMed: https://pubmed.ncbi.nlm.nih.gov/33529167/
- Zhao Y, Kilian C, Turner JE, Bosurgi L, Roedl K, et al. Clonal expansion and activation of tissue-resident memory-like Th17 cells expressing GM-CSF in the lungs of severe COVID-19 patients. Sci Immunol. 2021; 6: eabf6692. PubMed: https://pubmed.ncbi.nlm.nih.gov/33622974/
- Karki R, Sharma BR, Tuladhar S, Williams EP, Zalduondo L, et al. COVID-19 cytokines and the hyperactive immune response: Synergism of TNF-α and IFN-γ in triggering inflammation, tissue damage, and death. Cell. 2021; 184: 149-168.
- Hossain MK, Hassanzadeganroudsari M, Apostolopoulos V. The emergence of new strains of SARS-CoV-2. What does it mean for COVID-19 vaccines? Expert Rev Vaccines. 2021; 1-4. PubMed: https://pubmed.ncbi.nlm.nih.gov/33896316/
- Lavine JS, Bjornstad ON, Antia R. Immunological characteristics govern the transition of COVID-19 to endemicity. Science. 2021; 371: 741-745. PubMed: https://pubmed.ncbi.nlm.nih.gov/33436525/
- Altmann DM, Boyton RJ, Beale R. Immunity to SARS-CoV-2 variants of concern. Science. 2021; 371: 1103-1104. PubMed: https://pubmed.ncbi.nlm.nih.gov/33707254/
- Yuen KS, Ye ZW, Fung SY, Chan CP, Jin DY. SARS-CoV-2 and COVID-19: The most important research questions. Cell Biosci. 2020; 10: 40.
- Saad-Roy CM, Wagner CE, Baker RE, Morris SE, Farrar J, et al. Immune life history, vaccination, and the dynamics of SARS-CoV-2 over the next 5 years. Science. 2020; 370: 811-818. PubMed: https://pubmed.ncbi.nlm.nih.gov/32958581/
- Lythgoe KA, Hall M, Ferretti L, de Cesare M, MacIntyre-Cockett G, et al. SARS-CoV-2 within-host diversity and transmission. Science. 2021; 372: eabg0821. PubMed: https://pubmed.ncbi.nlm.nih.gov/33688063/
- Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 2020; 21: 1336-1345. PubMed: https://pubmed.ncbi.nlm.nih.gov/32887977/
- Abu-Raddad LJ, Chemaitelly H, Adeel A. Butt AA. Effectiveness of the BNT162b2 Covid-19 Vaccine against the B.1.1.7 and B.1.351 Variants. N Engl J Med. 2021; 385: 187-189. PubMed: https://pubmed.ncbi.nlm.nih.gov/33951357/
- Gómez CE, Perdiguero B, Esteban M. Emerging SARS-CoV-2 Variants and Impact in Global Vaccination Programs against SARS-CoV-2/COVID-19. Vaccines (Basel). 2021; 9: 243. PubMed: https://pubmed.ncbi.nlm.nih.gov/33799505/
- Benefits of Getting a COVID-19 Vaccine. Center for Disease control and Prevention. 2021. https://www.cdc.gov/coronavirus/2019-ncov/vaccines/vaccine-benefits.html
- Mysterious Ailment, Mysterious Relief: Vaccines Help Some COVID Long-Haulers. 2021. https://www.npr.org/sections/health-shots/2021/03/31/982799452/mysterious-ailment-mysterious-relief-vaccines-help-some-covid-long-haulers
- Sironi M, Hasnain SE, Rosenthal B, Phan T, Luciani F, et al. SARS-CoV-2 and COVID-19: A genetic, epidemiological, and evolutionary perspective. Infect Genet Evol. 2020; 84: 104384. PubMed: https://pubmed.ncbi.nlm.nih.gov/32473976/
- Plante JA, Mitchell BM, Plante KS, Debbink K, Weaver SC, et al. The variant gambit: COVID-19's next move. Cell Host Microbe. 2021; 29: 508-515. PubMed: https://pubmed.ncbi.nlm.nih.gov/33789086/
- Cooper V. The Coronavirus Variants Don’t Seem to Be Highly Variable So Far: SARS-CoV-2 may be settling into a limited set of mutations. Scientific American. 2021. https://www.scientificamerican.com/article/the-coronavirus-variants-dont-seem-to-be-highly-variable-so-far